Recent advances in flow cytometry enable simultaneous single-cell measurement of 30+ surface and intracellular proteins. In a single experiment we can now measure enough markers to identify and compare functional immune activities across nearly all cell types in the human hematopoietic lineage. However, practical approaches to analyze and visualize data at this scale are only now becoming available. SPADE, described in Qiu et al., Nature Biotechnology 2011 and first used in Bendall et al. Science 2011 , is a novel algorithm that organizes cells into hierarchies of related phenotypes, or "trees", that facilitate the visualization of developmental lineages, identification of rare cell types, and comparison of functional markers across stimuli.
CytoSPADE is a robust, modular and performant implementation of Qiu et al.'s SPADE algorithm, including a rich GUI implemented as a plugin for the Cytoscape Network Visualization platform. Explore these pages to get started with SPADE, and to learn more about the SPADE algorithm, the CytoSPADE implementation and other advanced tools for working with flow cytometry data, e.g., Gemstone, developed elsewhere.
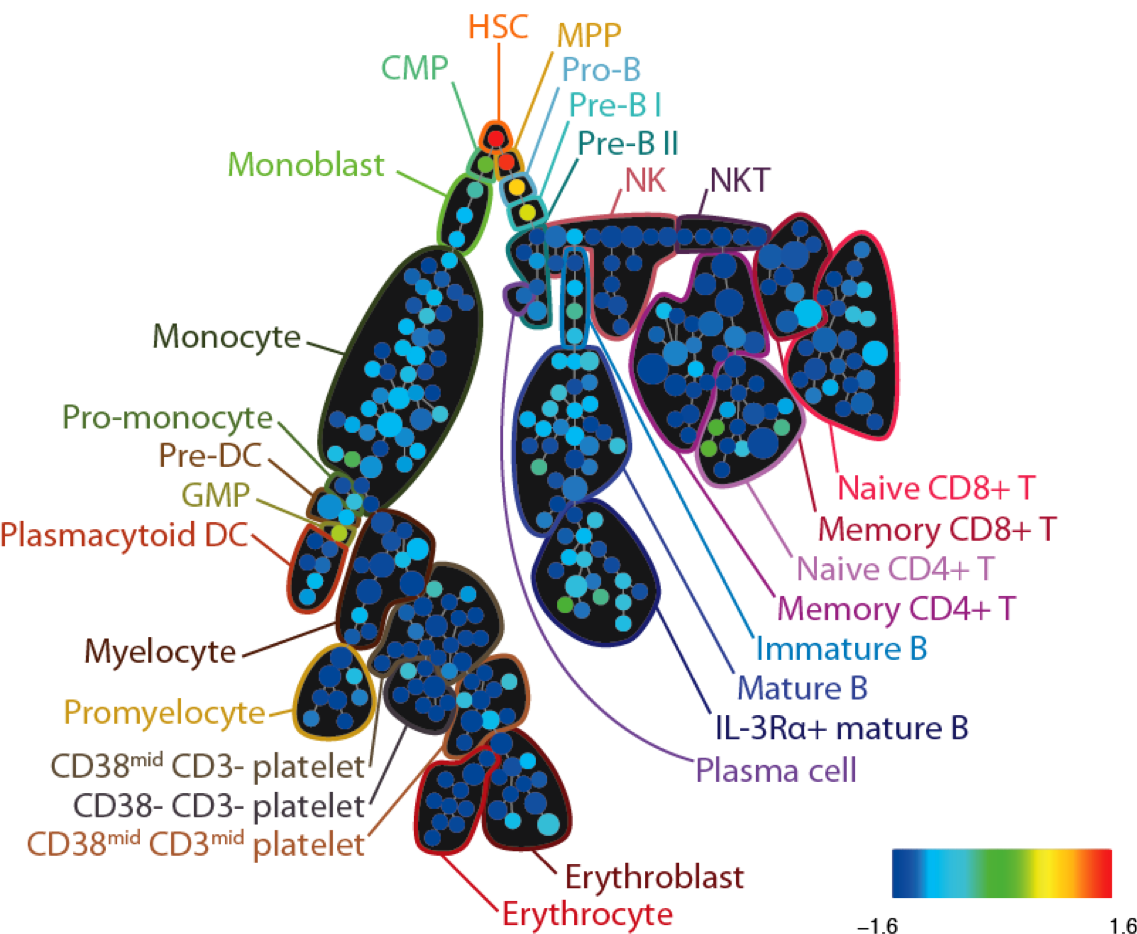
SPADE tree annotated with CD34 expression in healthy human bone marrow (PBMC) samples analyzed via mass cytometry. Adapted from Bendall et al. Science 2011
